The KRAS gene encodes the KRAS protein, a small GTPase that serves as a key molecule in cellular signal transduction, regulating critical physiological processes such as cell proliferation, differentiation, and survival. Among human cancers, KRAS mutations are one of the most common oncogenic drivers, with the KRAS[G12D] mutation (glycine at position 12 replaced by aspartic acid) being particularly prominent. This mutation causes the KRAS protein to remain persistently activated, triggering uncontrolled cell growth and division, and is closely associated with the development of various malignant tumors.
Statistics show that the KRAS[G12D] mutation is present in:
- Over 50% of pancreatic ductal adenocarcinomas
- Approximately 35% of colorectal cancers
- About 15% of non-small cell lung cancers
Although KRAS has long been considered an "undruggable" target, recent years have seen major breakthroughs in KRAS-targeted therapy with the approval of KRAS G12C-specific inhibitors such as Sotorasib (AMG-510) and Adagrasib (MRTX849). However, targeted drugs for the KRAS[G12D] mutation are still in the development stage, representing a significant unmet clinical need.
![Schematic_Diagram_of_KRAS[G12D]_Protein_Structure_and_Function.png Schematic_Diagram_of_KRAS[G12D]_Protein_Structure_and_Function.png](/uploads/image/20250613/Schematic_Diagram_of_KRAS[G12D]_Protein_Structure_and_Function.png)
Figure 1. Schematic Diagram of KRAS[G12D] Protein Structure and Function
Three Strategies for KRAS[G12D] Drug Development:
* Blocking upstream activation signals: Inhibiting the interaction between KRAS[G12D] and the guanine nucleotide exchange factor SOS1 to prevent GDP/GTP exchange, keeping KRAS in an inactive state.
* Directly inhibiting GTP binding: Preventing KRAS[G12D] from binding to GTP and blocking its activation process.
* Disrupting downstream signaling: Inhibiting the binding of KRAS[G12D] to effector proteins (e.g., RAF1) to block MAPK pathway activation.
To accelerate drug development for these strategies, high-quality, highly simulated in vitro screening tools are particularly important. Most screening kits currently on the market use conventional KRAS[G12D] protein, while VKEY-BIO's innovative GDP-loaded KRAS[G12D] protein-based assay kits more accurately mimic the physiological environment, providing more reliable data support for drug screening.
Using its self-developed KeyTec® TR-FRET (Time-Resolved Fluorescence Resonance Energy Transfer) technology, VKEY-BIO has launched a series of KRAS[G12D] protein interaction assay kits, including:
- GDP-loaded KRAS[G12D]/SOS1 Binding Assay Kit (Cat. No.: A1090001)
- GDP-loaded KRAS[G12D]/RAF1 Binding Assay Kit (Cat. No.: A1090002)
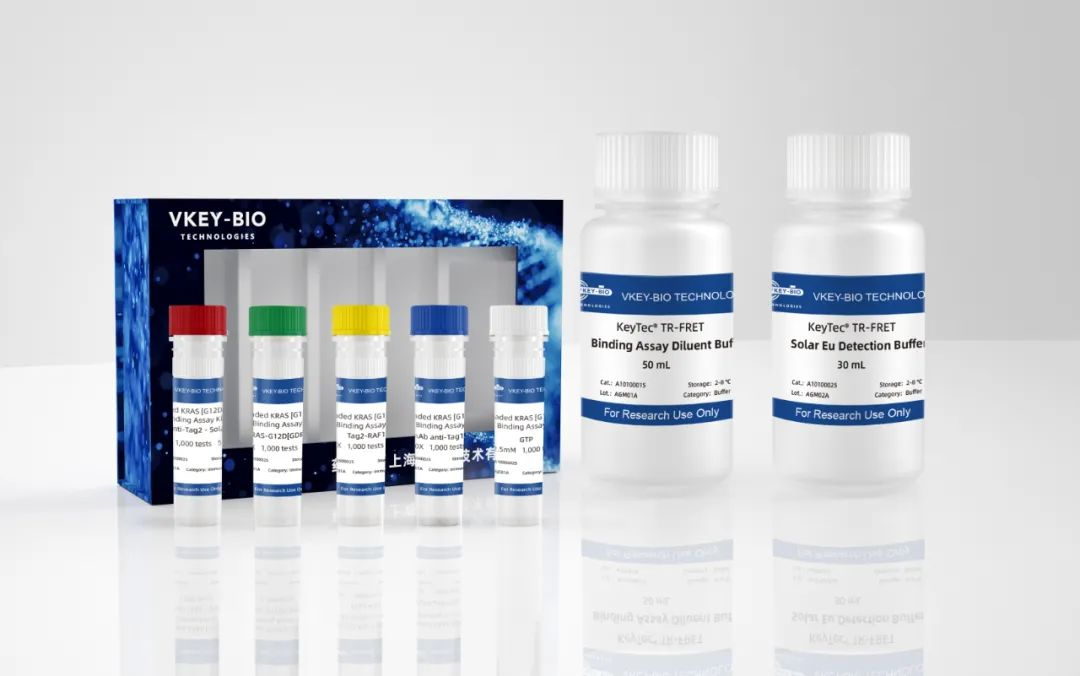
Figure 2. Product Packaging Schematic
These kits utilize GDP-loaded KRAS[G12D] mutant protein to more accurately simulate the natural state of KRAS protein in cells, providing physiologically relevant experimental conditions for drug screening.
The Solar Eu (donor)-labeled anti-Tag2 antibody specifically recognizes Tag2-labeled SOS1 protein, while the LA (acceptor)-labeled anti-Tag1 antibody specifically recognizes Tag1-labeled KRAS[G12D] protein. When SOS1 binds to KRAS[G12D], the two labeled antibodies come into proximity, resulting in fluorescence resonance energy transfer upon excitation by an external light source. By measuring the signal intensity at 665 nm, the degree of protein interaction can be quantified. Compounds that block this interaction will reduce the TR-FRET signal, enabling high-throughput screening.

Figure 3. Schematic Diagram of KeyTec® TR-FRET GDP-loaded KRAS[G12D]/SOS1 Binding Assay Principle
Outstanding Performance: High Sensitivity and Stability
The kit demonstrates excellent performance in multiple tests.
![Performance_Chart_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]SOS1_Kit.png Performance_Chart_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]SOS1_Kit.png](/uploads/image/20250613/Performance_Chart_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]SOS1_Kit.png)
Figure 4. Performance Chart of KeyTec® TR-FRET GDP-loaded KRAS[G12D]/SOS1 Kit
This kit employs a TR-FRET design similar to the SOS1 kit but tailored for RAF1: Solar Eu-labeled anti-Tag2 antibody recognizes Tag2-RAF1, while LA-labeled anti-Tag1 antibody recognizes Tag1-KRAS[G12D]. When KRAS[G12D] binds to RAF1, energy transfer occurs, and the binding strength can be quantified by measuring 665 nm fluorescence. Potential inhibitors will reduce the signal.
![Schematic_Diagram_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]RAF1_Binding_Assay_Principle.png Schematic_Diagram_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]RAF1_Binding_Assay_Principle.png](/uploads/image/20250613/Schematic_Diagram_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]RAF1_Binding_Assay_Principle.png)
Figure 5. Schematic Diagram of KeyTec® TR-FRET GDP-loaded KRAS[G12D]/RAF1 Binding Assay Principle
Excellent Screening Performance:
![Performance_Chart_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]RAF1_Kit.png Performance_Chart_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]RAF1_Kit.png](/uploads/image/20250613/Performance_Chart_of_KeyTec®_TR-FRET_GDP-loaded_KRAS[G12D]RAF1_Kit.png)
Figure 6. Performance Chart of KeyTec® TR-FRET GDP-loaded KRAS[G12D]/RAF1 Kit
Comparison with Traditional KRAS Binding Assay Kits, KeyTec® TR-FRET series products offer significant advantages:
Advantage | Description | User Value |
Simple and Fast | Three-step operation: "add sample-incubate-read plate" for rapid detection | Greatly improves screening efficiency, suitable for high-throughput screening |
High Accuracy | Uses GDP-loaded KRAS[G12D] protein, closer to physiological state | More physiologically relevant screening results, reducing false positives |
High Signal-to-Noise Ratio | Patented Solar Eu/LA fluorescent pair for higher signal-to-noise ratio | More reliable data, easier interpretation of results |
Excellent Reproducibility | Strict quality control | Stable and reliable data, supporting long-term research |
- GDP Pre-loading Technology: Simulates the natural state of KRAS protein in cells, improving physiological relevance in screening.
- Dual-Tag System: Tag1-KRAS[G12D] and Tag2-SOS1/RAF1 ensure detection specificity.
- Pre-mixed Antibody Design: Reduces operational steps and minimizes human error.
- Optimized Buffer System: Enhances protein stability and ensures assay reproducibility.
- Small-molecule inhibitor screening: High-throughput screening for compounds that block KRAS-SOS1/RAF1 interactions.
- Antibody drug development: Evaluating the blocking effects of anti-KRAS or anti-SOS1/RAF1 antibodies.
- Mechanism studies: Investigating the impact of KRAS mutations on protein interactions.
- Drug optimization: Assessing the effects of compound structural modifications on activity.
Product Specifications
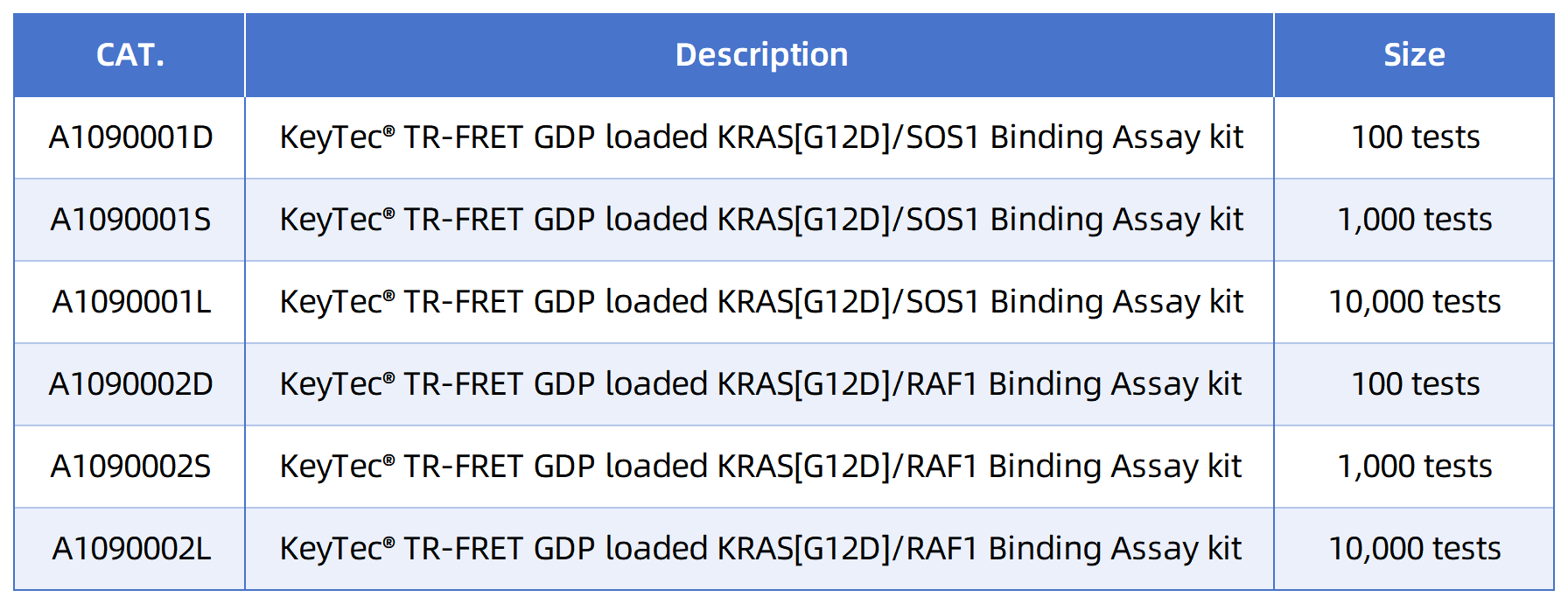
References
1. Mao Z, Xiao H, Shen P, et al. KRAS (G12D) can be targeted by potent inhibitors via formation of salt bridge[J]. Cell discovery, 2022, 8(1): 5.
2. Farren M R, Roman V, Gallion A, et al. INCB161734: A novel, potent, and orally bioavailable KRAS G12D selective inhibitor demonstrates antitumor activity in KRAS G12D mutant tumors[J]. Cancer Research, 2024, 84(6_Supplement): 5900-5900.
3. Zheng Q, Zhang Z, Guiley K Z, et al. Strain-release alkylation of Asp12 enables mutant selective targeting of K-Ras-G12D[J]. Nature Chemical Biology, 2024: 1-9.
4. Vatansever S, Erman B, Gümüş Z H. Oncogenic G12D mutation alters local conformations and dynamics of K-Ras[J]. Scientific reports, 2019, 9(1): 11730.
5. Bryant, K. L., et al. (2014). "KRAS: Feeding pancreatic cancer proliferation." Trends in Biochemical Sciences, 39(2), 91-100.
6. TCGA Network. (2012). "Comprehensive molecular characterization of human colon and rectal cancer." Nature, 487(7407), 330-337.
7. TCGA Network. (2014). "Comprehensive molecular profiling of lung adenocarcinoma." Nature, 511(7511), 543-550.